Research in our lab focuses on evolutionary and developmental questions of plant biology. Currently our primary focus is on functional genomics of phytochrome signaling. Other long-term interests in the lab are the evolutionary biology and physiology of plant polyploids.
Current Projects
Updated: November 2023
Functional Genomics of Phytochrome Responses
Throughout development, signal transduction plays an important role in determining cell fate and establishing a developmental program so that a single cell can develop into a multicellular organism. Signal transduction uses signaling pathways, which are circuits that convey messages between cells and their environment. The response to external signals is context dependent, meaning the output depends not only upon the cell type in which signals are transduced, but also upon the stage of development of the organism, other interacting signaling molecules, and outside environmental signals.
We focus our studies on the interaction between light perception and growth hormones, which implement the perceived signals and translate them into appropriate gene responses. Specifically, we study the function of several lesser-known genes in the phytochrome family of red-light photoreceptors and their interaction with downstream signal transduction molecules. We use integrative approaches, combining the power of whole genome analysis, such as RNA-seq and proteomics, with classical physiological approaches to understand which genes are involved in specific signaling responses. We are also interested in better understanding how light receptor genes interact with the rest of the genome using bioinformatic gene network analysis.
Evolutionary Biology of Genome Structure (Polyploidy)
Our lab has a long-standing interest in genome structure and its role in plant evolution. Plant genomes are remarkably dynamic and structural change is tolerated in plants much more readily than in animals. One of the most frequent structural changes in plant genomes involves the duplication of the entire chromosome complement leading to a state known as polyploidy. Two types of polyploids are distinguished: allopolyploids (hybrids with duplicated genomes of two different species) and autopolyploids (duplicated genomes of one species).
Because polyploid genomes carry more loci of every gene than diploids do, it has been suggested that they are more adaptive to their environment and are superior to diploids in their ability to evolve. Despite much recent work, there is still surprisingly little known about direct connections between polyploidy, polyploidy-induced genetic changes, and corresponding physiological responses that directly affect the polyploid’s phenotype under natural conditions (discussed in more detail at Madlung 2013). Using the model genus Arabidopsis we study physiological changes and their underlying molecular mechanisms in polyploids that could lead to speciation, adaptation, and genome evolution.
Educational research
I am interested in the question of how best to introduce undergraduate students to genome-scale data analysis. To this end I have developed various lab modules that I have tested over the years. The results of two such studies were reported in CBE Life Science Education (2011), and PLoS Computational Biology (2018).
Summaries of some of our publications (see full publication list)
|
Balderrama, D., Barnwell, S., Carlson, K.D., Salido, E., Guevara, R., Nguyen, C., and Madlung, A. (2023) Plant Physiology Phytochrome F mediates red lighy responsiveness additively with phytochromes B1 and B2 in tomato https://doi.org/10.1093/plphys/kiad028 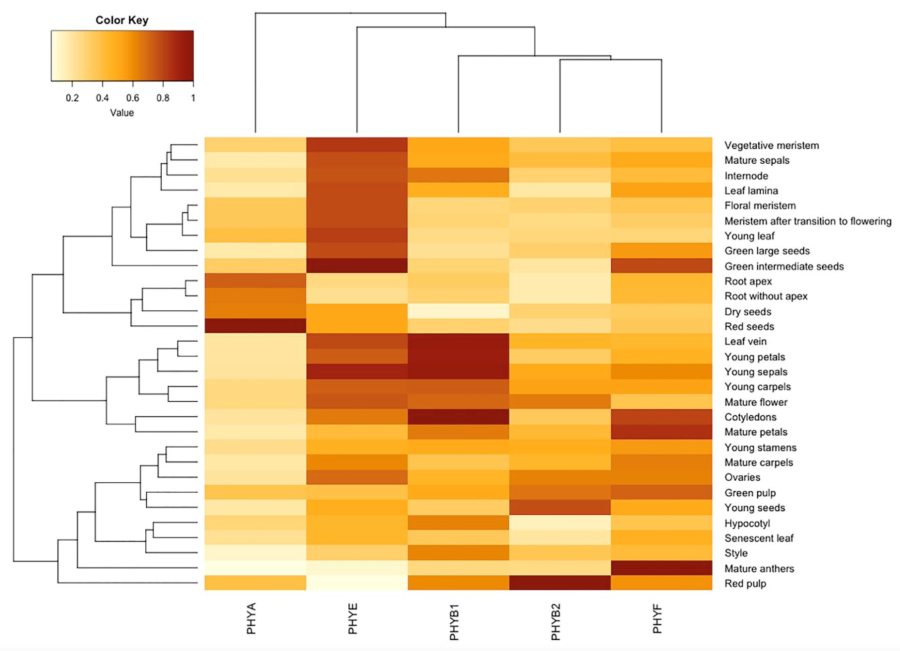
This paper describes the creation of a new mutant in PHYTOCHROME F in tomato and its phenotypic characterization. |
|
Orugante, V, Toegelova, H, Pečinka, A., Madlung, A. and Schneeberger, K. Rapid large-scale genomic introgression in Arabidopsis suecica via an autoallohexaploid bridge (2022) Genetics, 223 (2), iyac132, https://doi.org/10.1093/genetics/iyac132 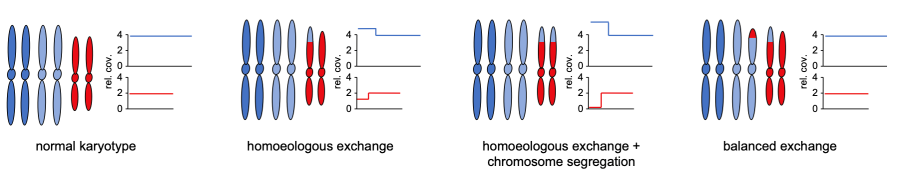
This paper describes the genomic changes that occur in a natural allohexaploid Arabidopsis population over multiple generations and suggests that allohexaploidy serves as a bridge to facilitate rapid gene flow between two species. |
|
Carlson, KD, Bhogale, S, Anderson, D, Zaragoza-Mendoza, A, Madlung, A. Subfunctionalization of phytochrome B1/B2 leads to differential auxin and photosynthetic responses. (2020) Plant Direct 4(2): e00205. (Full text) 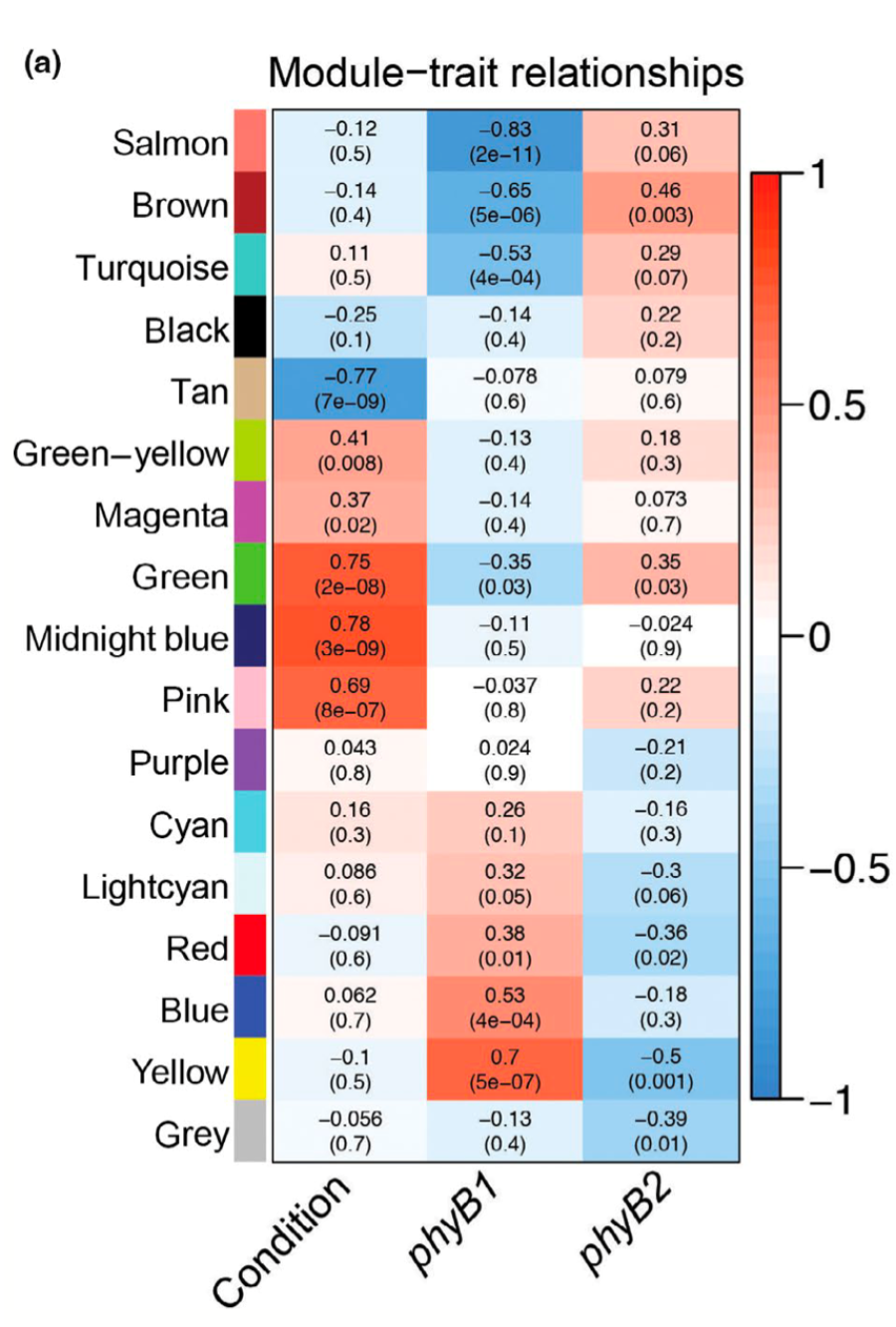
In this paper, we use transcriptional network analysis followed by physiological verification to show that tomato PHYTOCHROME B1 and PHYTOCHROME B2 diversified after duplication with respect to their involvement in auxin responses and photosynthesis. |
|
Carlson, KD, Bhogale, S, Anderson, D, Tomanek, L, Madlung, A. Phytochrome A regulates carbon flux in dark grown tomato seedlings. (2019) Frontiers in Plant Science 10(152) (Full text) 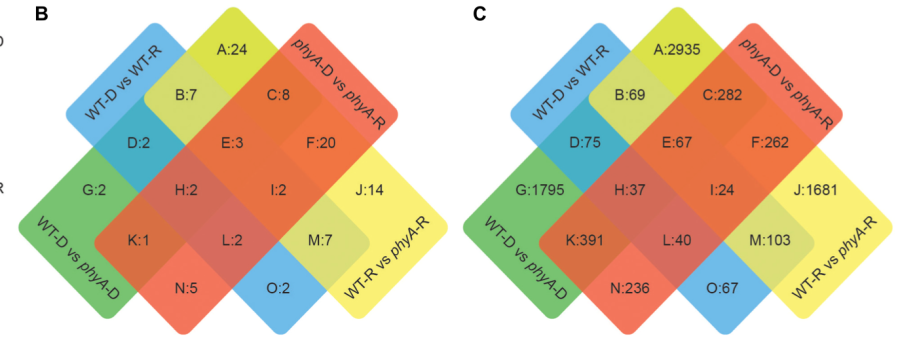
Using whole genome transcriptional network analysis, in this study we found that tomato PHYTOCHROME A participates in the regulation of carbon flux through major primary metabolic pathways, including glycolysis and the TCA cycle. |
|
Carlson, KD, Fernandez-Pozo, N, Bombarely, A, Pisupati, R, Mueller, LM, and Madlung, A: Natural variation in stress response gene activity in the allopolyploid Arabidopsis suecica (2017) BMC Genomics 18:653 (Full text) 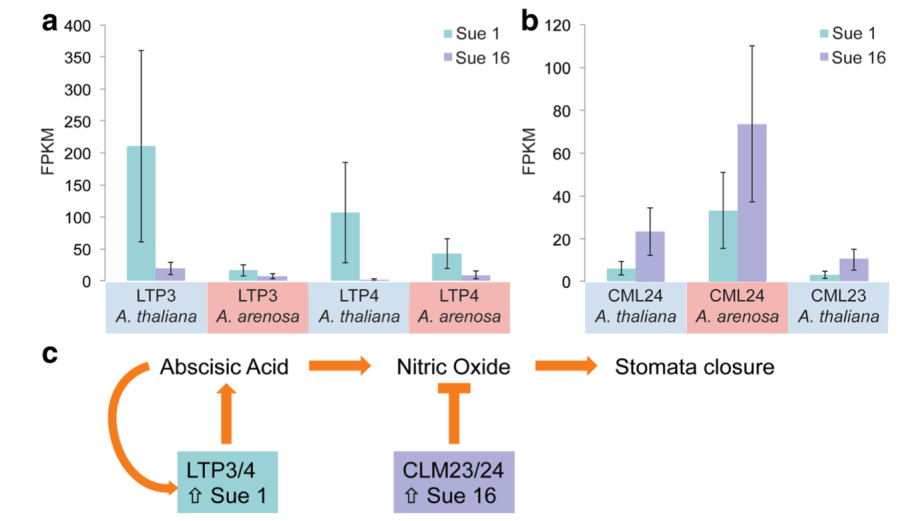
In this paper we describe how differentially expressed homoeologous genes in two different accessions of an allopolyploid lead to variation in transpiration and stomatal regulation, suggesting allopolyploidy as a mechanism in differential environmental adaptation. |
|
Solhaug, EM., Ihinger, J, Jost, M, Gamboa, V, Marchant, B, Bradford, D, Doerge, RW, Tyagi, A, Replogle, A, Madlung, A: Environmental regulation of heterosis in allopolyploid Arabidopsis suecica. (2016) Plant Physiology 170: 2251-2263, doi:10.1104/pp.16.00052 (Full text) 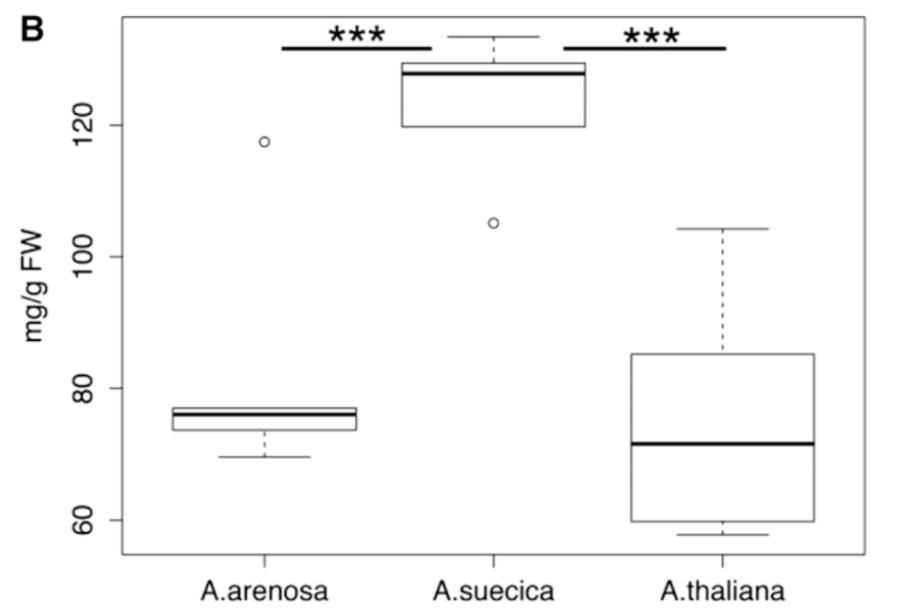
In this paper we show that the magnitude of hybrid vigor of photosynthetic activity and starch production in allopolyploids of Arabidopsis is dependent on the environmental conditions. |
|
Matsushita, SC, Tyagi, AP, Thornton, GM, Pires, JC, Madlung, A: Allopolyploidization lays the foundation for evolution of distinct populations: evidence from analysis of synthetic Arabidopsis allohexaploids. (2012) Genetics, 191, 535-547 (Abstract) 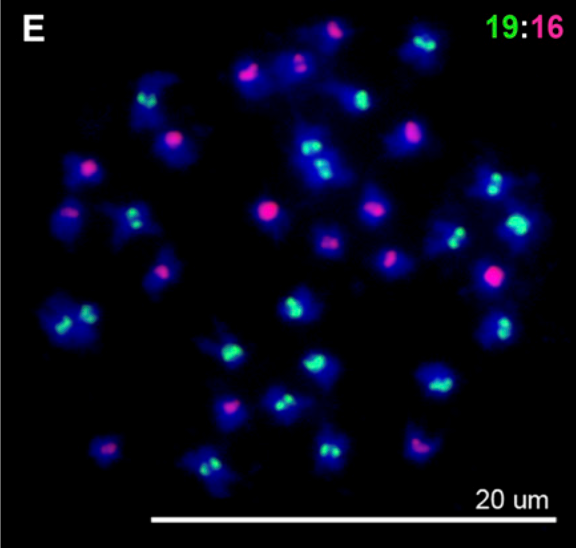
In this study we showed cytologically that allohexaploidy leads to chromosomal variation and somatic mosaicism. This variability might be underlying the wide-ranging phenotypic variability in allohexaploids. This study provided our first indication of hexaploidy as a mechanism for fast-track diversification. |
|
McCullough, E, Wright, KM, Alvarez, A, Clark, CP, Rickoll, WL, Madlung, A: Photoperiod-dependent floral reversion in the natural allopolyploid Arabidopsis suecica (2010) New Phytologist, 186, 239-250 (Abstract) 
In this paper we showed that allopolyploidy in Arabidopsis affects floral morphology in a photoperiod-dependent manner and coincides with the competing expression of floral promoter and floral repressors in the abnormal floral tissues. |
Getting involved
Currently we are mostly focusing on phytochrome work using tomato as a model. We are creating and characterizing additional mutants with the aim to better understand how all phytochrome genes interact with each other during morphogenesis and development of the plant. We are also interested in better understanding the influence the environment has on the diversity of the response to light cues.
All projects in the lab lend themselves well to independent research in genetic, molecular, and integrative biology. If you are interested in joining the lab, contact me and I can suggest a number of specific projects to you. A complete list as well as links to our published work are on our publications page.
See Also:
Contact Information
Andreas Madlung
Office: Thompson 223F
Lab: Thompson 217
253.879.2712
253.879.2764
amadlung@pugetsound.edu


